The plasma cell cancer multiple myeloma (MM) varies significantly in genomic characteristics, response to therapy, and long-term prognosis. Recent studies have shown the complexity of biological systems associated with poor outcomes in MM. One hallmark of cancer pathogenesis, applicable to MM, is the dysregulation of apoptotic cell death. Understanding the relationship between known drivers of MM and critical pathways in the apoptosis gene network is key towards identifying novel therapeutic targets.
Recently, there has been a surge of interest in developing network-based approaches to study gene interactions and use machine learning tools to further elucidate useful biological factors. It is well-known that genes operate as network systems; however, interpretable computational methods are still limited. In this work, we combine a set of network analysis tools to provide novel biological insights into how the apoptosis network is dysregulated in MM. We identify a set of genes associated with poor outcomes in MM using both univariate and network-based approaches.
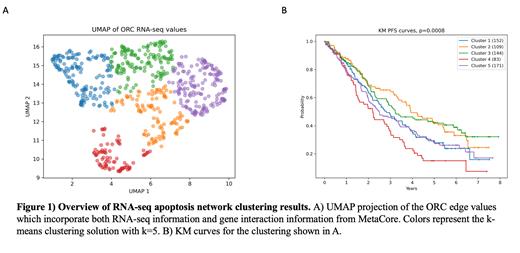
Genomic and clinical data, including RNA-sequencing (RNA-seq) and copy number alteration (CNA), for 659 subjects were obtained from the Multiple Myeloma Research Foundation's CoMMpass (IA19) database. The apoptosis gene set is defined using the apoptosis hallmark pathway provided by the gene set enrichment analysis tool. The interactions between the given list of genes are provided via MetaCore. Univariate gene modeling is done using a Cox proportional hazards model. To determine the relationship between genes, we used Ollivier-Ricci curvature (ORC), a measure of network robustness. The ORC value between two genes incorporates information about both the two genes in question and their local neighborhoods, a concept critical for understanding the role a neighborhood of genes has on a given connection. Once edge information is computed, we used a uniform manifold approximation and projection (UMAP) to reduce the complexity of the dataset. The resulting projection is clustered using k-means clustering, and survival analysis of the given clusters is done using the Kaplan-Meier (KM) method.
To understand which features are most informative, we used a random forest to predict the cluster labels using the RNA-seq data. After five-fold cross-validation, feature importance was computed using random forest's permutation importance methodology. All statistical significance testing was corrected for multiple comparisons via the Benjamini-Hochberg false discovery rate method with an alpha set to 0.05.
Using RNA-seq data of genes associated with apoptosis, we identified nineteen apoptosis genes associated with poor prognosis in MM. Some of these genes, such as ANXA1 and WEE1 are known to have an effect in MM, while others, such as AVPR1A have yet to be studied extensively for their role in MM. When examining CNA data associated with apoptosis, a similar pattern emerged - nineteen genes were associated with poor prognosis. However, between the two key gene sets found by RNA-seq and CNA, only FAS appeared in both lists. FAS, Fas Cell Surface Death Receptor, is critical for triggering apoptosis. Half of the 37 genes identified using RNA-seq and CNA data are directly connected to the TP53 gene. While TP53 mutations are not a marker of the presence of MM itself, TP53 mutations have been shown to be associated with poorer outcomes.
After reducing the complexity of the Ollivier-Ricci curvature analysis of the RNA-seq apoptosis network using UMAPs, the data revealed five clusters with KM curves for progression-free survival were significant (p<0.001). To identify the genes most impactful for separating these clusters, we trained a random forest model on the RNA-seq - note, it was the edge robustness values that were clustered - and evaluated its performance for predicted class membership. The model had an overall accuracy of approximately 78% with F1 scores for each class ranging from 61% to 87%. The top three features associated with the model include CTNNB1, RELA, and LEF1. Of note, CTNNB1 and RELA are directly interacted with the TP53 gene. Because these three genes discriminate between all five clusters, their expression levels may play a mediating effect on MM outcomes. Further investigation with external datasets is needed to validate these results.
Disclosures
Usmani:GSK: Membership on an entity's Board of Directors or advisory committees, Research Funding; Gilead Sciences: Membership on an entity's Board of Directors or advisory committees, Research Funding; Genentech: Membership on an entity's Board of Directors or advisory committees; EdoPharma: Membership on an entity's Board of Directors or advisory committees; K36 Therapeutics: Membership on an entity's Board of Directors or advisory committees; Janssen: Membership on an entity's Board of Directors or advisory committees, Research Funding; Moderna: Membership on an entity's Board of Directors or advisory committees; Novartis: Membership on an entity's Board of Directors or advisory committees; Oncopeptides: Membership on an entity's Board of Directors or advisory committees; Sanofi: Membership on an entity's Board of Directors or advisory committees, Research Funding; Seattle Genetics: Membership on an entity's Board of Directors or advisory committees, Research Funding; SecuraBio: Membership on an entity's Board of Directors or advisory committees; SkylineDX: Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda: Membership on an entity's Board of Directors or advisory committees, Research Funding; TeneoBio: Membership on an entity's Board of Directors or advisory committees; Array Biopharma: Research Funding; Merck: Research Funding; Pharmacyclics: Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Bristol Meyer Squibb: Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Membership on an entity's Board of Directors or advisory committees, Research Funding; Abbvie: Membership on an entity's Board of Directors or advisory committees, Research Funding.